Nuclear poro – Wikipedia, the free encyclopedia

The Nuclear pores [ first ] The Nuclear pore complexes ( NPC In English) they are large protein groupings, which cross the nuclear envelope (EN), the double membrane that surrounds the cellular nucleus of the eukaryotes.
Each pore is a supra-macromolecular set, composed of multiple copies of 30 different proteins, giving a total of 500-800 nucleoporins per pore with an approximate molecular mass of 100 megadaltons (MDA).
Its composition and size can vary between eukaryotes, but its structure and general function seem to be preserved in different species.
There are about 4000 nuclear pore complexes, in the nuclear wrapping of the vertebrate cell, but its number varies depending on the cell type.
The nuclear pore complex (CPN) is the door of the nucleus and all the molecules must cross it to travel from the nucleus to the cytoplasm and vice versa.
The in and the CPN regulate transport and are also relevant regulators of the organization of chromatin and the expression of the genes.
Characteristics [ To edit ]
Nuclear pores are huge active macromolecular complexes and not simple passive holes in the nuclear envelope.
The concept of nuclear pore complex (CPN) ( Nuclear Pore Complex NPC in English) arose in the last quarter of the century XX , with the arrival of ultrastructural and biochemical study methods capable of solving these structures at the molecular level. [ 2 ]
Number [ To edit ]

The number and density of the nuclear pores (CPN complexes) varies between the different types of cells and within the life cycle of each of them. [ 3 ]
In vertebrates you can take an estimate of 11 pores per square micrometer (µm²) as average, in its nuclear wrap (EN). The extrapolation for the entire surface of the nucleus wrap, would give a total of 2000-4000 CPN complexes per nucleus. [ 4 ]
The yeasts, on average barely exceed 200 complexes for each µm² of its en. [ 5 ]
The number of nuclear pores (CPN) increases during the cell cycle and in response to hormonal stimulation. In Xenopus oocyte, the total number of NPC complexes increases with the maturation stadium, of 1.2 × 10 7 In Stage II until 3 × 10 7 In stadium VI. [ 6 ]
The cells that proliferate, such as embryos or tumors, have a high density of nuclear pores. [ 3 ]
Structure [ To edit ]
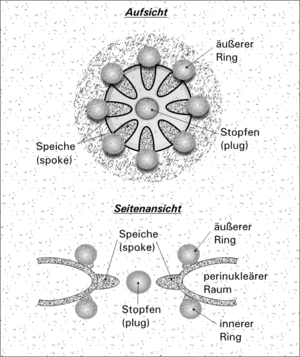
Nuclear pores are protein structures, embedded in the double membrane of the nuclear envelope. The nuclear pore (CPN) complex ring consists of two components:
- The external component of the CPN
It is located in the perinuclear cistern, in intimate association with the internal nuclear membranes and external nuclear that limit the pore.
- The internal component of the CPN
It is the central structure of the CPN with a schematic appearance of ‘cart wheel’, ‘Rosquilla’ or ‘Octagonal wheel’, is made up of nucleoporine proteins with a total molecular weight of 50-60 megadaltons (MDA). It consists of eight radios, arranged around the central channel, which serves as a duct for the transport of macromolecules. [ 7 ] [ 8 ] [ 9 ]
| Body | Size | Peso | Investigator |
|---|---|---|---|
| Yeast | 102-128 nm | 60-66 MDa | Fahrenkrog [ ten ] 2011 |
| Xenopus | 120-150 nm | 110-125 MDa | Cohen [ 11 ] 2012 |
| Human | ~140 nm | ~110 MDa | Lin [ 2 ] 2019 |
The Pore complex measures about 100-150 nm in diameter, with about 40 nm in useful internal diameter, and 50-70 nm high. [ 4 ]
The internal and nuclear nuclear membranes of the nuclear envelope are joined by the Membrane domain Where the CPN is housed. The external rings or cytoplasmic ring of the CPN, and the internal ring or nuclear ring, are located in the perinuclear space in intimate association with the nuclear membrane that limits the pore. [ 7 ]
The CPN is a supra-macromolecular set, composed of multiple copies of 30 different families, with a total of 456 nucleoporins (NUPS).
Nucleoporinas [ To edit ]

Proteins that form nuclear pore complexes are known as nucleoporins (NUPS).
The CPN is a supra-macromolecular set, composed of multiple copies of 30 different nucleoporins, being ~ 500 nupts in total, 800 in mammals. [ twelfth ]
About half of the nucleoporins commonly contain an alpha solenoid or beta propeller tertiary structure, or in some cases both as separate protein domains. The other half shows typical structural characteristics of “native not folded” proteins, for example they are highly flexible proteins that lack an orderly secondary structure. [ 13 ] These disorderly proteins are FG nucleoporins, named for their amino acid sequence that contains several repetitions of the phenylalanine-glycine peptide [ 14 ]
The CPN nuclear pore complex ( NPC In English) it is the nucleus gate and all macromolecules must cross it to travel from the nucleus to the cytoplasm and vice versa. [ 5 ]
Nuclear pores allow the transport of water soluble molecules through the double membrane of the nuclear wrapping. This transport includes the RB and Ribosomal RNA movement from the nucleus to the cytoplasm, and the movement of proteins such as polymerases and sheets, in addition to carbohydrates, signal molecules and lipids towards the nucleus. [ 15 ]
It has been calculated that the CPN nuclear pore complex or ( NPC in English) can actively drive 1000 translocations per second in each complex.
Small molecules, such as metabolites and ions, pass freely through NPC. The diffusion of the largest molecules, is restricted by its sizes and superficial properties. Macromolecules nuclear import and export requires nuclear transport receptors ( NTR in English) that cross NPCs through the dissemination facilitated. [ twelfth ] [ 16 ]
Although small molecules pass by simple diffusion through pores, larger molecules can be recognized by specific signal sequences and then disseminated with the help of nucleoporins to or from the nucleus. This is known as the RAN cycle. Each of the eight protein subunits that surround the true pore (the external ring) projects a radio -shaped protein to the pore channel. The center of pore often seems to have a structure similar to a plug. It is not yet known whether this corresponds to a true cap or is simply loaded trapped during traffic.
Transportation through the nuclear pore complex [ To edit ]
Small particles (<50 kda) are capable of passing through the nuclear pore complex by passive diffusion. Larger particles can also pass through the large diameter of pore, but almost insignificant rates [ 17 ] The efficient step through the complex requires several protein factors. [ 18 ] The simplicity of transport by nuclear pores is facilitated by receptors in the FG domains called Carioferines , which are required for core-cytoplasmic transport of molecules greater than 40 kda. In the absence of these receptors, abbreviated KAPS, FG domains impose a physical barrier that prevents the passage of macromolecules through nuclear pore. [ 16 ] Carioferines, which can act as importins or exportations, are part of the importation of the importin -β that completely share a similar three -dimensional structure.
Three models have been suggested to explain the translocation mechanisms:
- Affinity gradients through the cap ( plug ) central.
- Browniana opening affinity.
- Selective phase.
Import of soluble proteins [ To edit ]
Any particle that carries a nuclear location signal (NLS) will be directed by the rapid and efficient transport through pore. Many nuclear location signs are known, they usually contain a sequence of amino acids preserved with basic waste such as Pkkkrkv. Any material with a nuclear location signal will be carried by imports to the nucleus.
The classic scheme for the importation of particles with a nuclear location signal begins first with the α-importine joining the sequence of the nuclear location signal, and acts as a bridge to unite β-importin. Then the load-βimportin-αimportin complex is directed towards nuclear pore and spreads through it. Once the complex is in the nucleus, Rangtp joins βimportin and displaces the complex. Then the Suceptibility protein of cell apoptosis (CAS), an export that is linked to RANGTP in the nucleus, separates the α-importing of the load. The nuclear location signal protein is found in this free way in the nucleoplasm. The βimportin-Rangtp and αimportin-CAS-RANGTP complexes spread back to the cytoplasm where GTPs are hydrolyzed to GDP leading to the release of βimportin and αimportin that is again available for a new importation of nuclear location signal.
Although the load passes through pore with the assistance of chaperone proteins, translocation through pore is not by itself dependent. However, the full import cycle needs the hydrolysis of 2 GTPS and therefore is dependent energy and has to be considered as active transport. The import cycle works thanks to the gradient of Rangtp Core-Cytoplasmic. This gradient arises from the exclusive nuclear location of Rangefs, proteins that change GDP to GTP in RAN molecules. Therefore there is a high rangtp concentration in the nucleus compared to the cytoplasm.
Protein export [ To edit ]
Some nuclear molecules need to be exported from the nucleus to the cytoplasm, such as ribosomal subunits and messenger arns. Therefore there is an export mechanism similar to import.
In the classic export scheme, proteins with a nuclear export sequence (NES) can be joined in the nucleus to form a heterotrophic complex with an export and RANGTP (for example CRM1 export). The complex can spread to the cytoplasm where the GTP is hydrolyzed and the nuclear export sequence protein is released. The CRM1-RANGDP complex spreads back to the nucleus where the GDP is changed to GTP by the Rangefs. This process is also energy dependent as it consumes GTP. Export with CRM1 can be inhibited by leptomycin C.
See also [ To edit ]
References [ To edit ]
- ↑ OMS,OPS (ed.). «Poro nuclear» . Descriptors in Health Sciences, Virtual Health Library .
- ↑ a b Lin D.H.; Hoelz A. (2019). «The Structure of the Nuclear Pore Complex (An Update)» . Annu Rev Biochem. (Revision) 88 : 725-783. PMID 30883195 . doi: 10.1146/annurev-biochem-062917-011901 . Retrieved on July 25, 2021 .

- ↑ a b Maeshima k .; yahata k .; sasaki y.; Nakatomi r .; tachibana T .; Hashikawa T .; Imamoto f .; Imamoto n. (2006). «Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins» . J Cell Sci 119 (21): 4442-4451 . Retrieved on July 27, 2021 .

- ↑ a b Megías M .; Molist P .; Pombal M.A. (2019). «4: The nucleus. Nuclear pores » . Plant and animal histology atlas. The cell .
- ↑ a b Geydan t.d.; Garzone-Coral c.; Fajardo C.; Spinel C. (2010). «Dynamics of the nuclear pore complex» . Acta biol. Colomb. (Scielo) 15 (1): 245-252.

- ↑ SELLÉS J.; Penrad-Mobayed M.; Guillaume C.; Fuger A.; Auvray L.; Faklaris O.; Montel F. (2017). «Nuclear pore complex plasticity during developmental process as revealed by super-resolution microscopy» . Scientific Reports 7 (14732 (2017)) . Retrieved on July 31, 2021 .

- ↑ a b Hoeijmakers J.H.J.; Schel J.H.N.; Wanka F. (1974). «Structure of the nuclear pore complex in mammalian cells: Two annular components» . Experimental Cell Research eighty seven (1): 195-206 . Retrieved on July 25, 2021 .
- ↑ Adam S.A. (2001). «The nuclear pore complex» . Genome Biology (Revision) 2 (reviews0007.1 (2001)) . Retrieved on July 26, 2021 .
- ↑ Hachiya n.; Sochocka m.; Brzecka A.; Shimizu t.; Gąsiorowski k.; Szczechowiak k.; Leszek J. (2020). «Nuclear Envelope and Nuclear Pore Complexes in Neurodegenerative Diseases—New Perspectives for Therapeutic Interventions» . Molecular Neurobiology (Review) (Springer) 58 : 983-995 . Retrieved on July 26, 2021 .

- ↑ Fahrenkrog B.; Stoffler D.; Aebi U. (2011). Nuclear Pore Complex Architecture and Functional Dynamics . Research Gate. Nuclear Export of Viral RNAs pp.95-117. doi: 10,1007/978-3-642-56597-7_5_5 .
- ↑ Cohen S.; Panté N. (2012). «2 NPC and Nuclear Transport». International Review of Cell and Molecular Biology .
- ↑ a b Cut U.; Jühlen r.; Anthony W. (2021). «Mitotic disassembly and reassembly of nuclear pore complexes» . Trends in Cell Bbiology (Revision) (in English) (Science Direct) . Retrieved on July 25, 2021 .
- ↑ Denning D, Patel S, Uversky V, Fink A, Rexach M (2003). «Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded» . Proc Natl Acad Sci U S A 100 (5): 2450-5. PMID 12604785 .
- ↑ Peters R (2006). «Introduction to nucleocytoplasmic transport: molecules and mechanisms» . Methods Mol Biol 322 : 235-58. PMID 16739728 . Filed from the original September 28, 2007.
- ↑ Colwell L.J.; Brenner M.P.; Ribbeck K. (2010). «Charge as a Selection Criterion for Translocation through the Nuclear Pore Complex.» . PLOS Computational Biology 6 (4)): E1000747. doi: 10.1371 / Journal.pcbi.1000747 . Retrieved on August 1, 2021 .

- ↑ a b LIM RY, Fahrenkrog B, et al. Nanomechanical Basis of Selective Gating by the Nuclear Pore Complex. Science. 2007 Oct 26. 318(5850):640-643. Available Last access November 22, 2007
- ↑ Rodríguez M, Dargemont C, Stutz F (2004). «Nuclear export of RNA». Biol Cell 96 (8): 639-55. PMID 15519698 .
- ↑ Reed R, Hurt E (2002). «A conserved mRNA export machinery coupled to pre-mRNA splicing». Cell 108 (4): 523-31. PMID 11909523 .


Recent Comments